Journal of Experimental Botany 62: 5131–5147 (2011)
Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination [W][OA]
*Department of Biological Sciences, Simon Fraser University, 9 8888, University Drive, Burnaby BC, V5A 1S6, Canada
Received 14 March 2011; Revised 22 May 2011; Accepted 13 June 2011
Advance Acess publication 21 June 2011
DOI 10.1093/jxb/err214
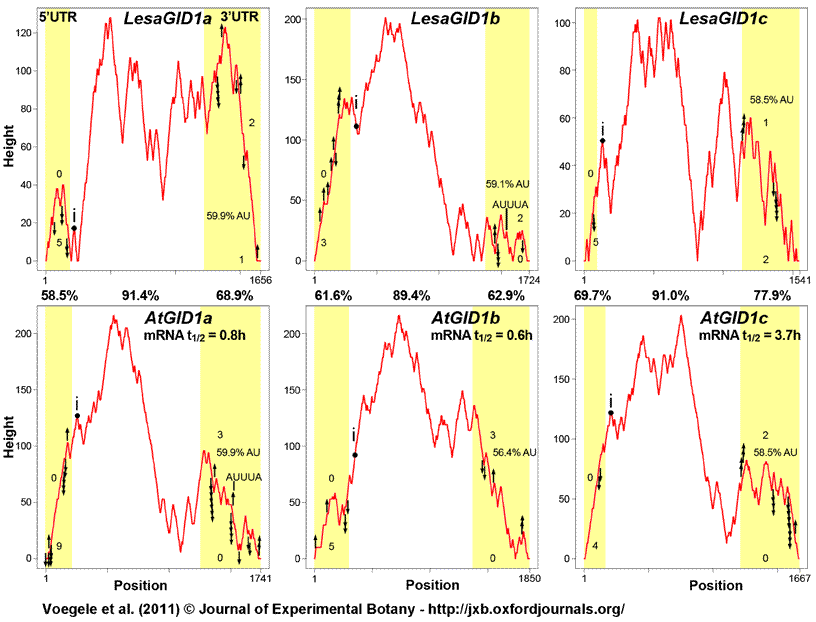
Supplemental Figure S3: Comparison of L. sativum and Arabidopsis GID1 transcript secondary structures and stability motifs in 5’ and 3’ UTRs (untranslated regions).
Secondary structures of L. sativum and Arabidopsis GID1 transcripts were predicted using the Vienna RNA Websuite tool RNAfold (Hofacker and Stadler, 2006) and were graphically represented using the mountain plot output, which plots secondary structures as height versus position. The height represents the number of bases which enclose a base at a peak or plateau position, which leads to loops being shown as plateaus and hairpins being represented as peaks. The positions of intron splice sites and different regulatory motifs are marked in each mountain plot diagram, as well as mRNA half lives (as determined in Arabidopsis cell cultures by Narsai et al., 2007) and AU contents in 3’ UTRs. Global UTR motifs associated with stabilizing and destabilizing of mRNAs as determined by Narsai et al. (2007) were marked using arrows pointing upwards (stabilizing motifs) or downwards (destabilizing motifs). The numbers of motifs related to the mRNA 5’-3’ decapping pathway as defined by Jiao et al. (2008) were indicated above (repressing mRNA uncapping) and underneath (enhancing mRNA uncapping) the mountain plot. Comparison of GID1 secondary structures and stability motifs in 5’ and 3’ UTRs suggests rapid regulation of transcript abundances.
| Article in PDF format (1.5 MB) Supplementary data file (6.5 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |