Journal of Experimental Botany 62: 5131–5147 (2011)
Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination [W][OA]
*Department of Biological Sciences, Simon Fraser University, 9 8888, University Drive, Burnaby BC, V5A 1S6, Canada
Received 14 March 2011; Revised 22 May 2011; Accepted 13 June 2011
Advance Acess publication 21 June 2011
DOI 10.1093/jxb/err214
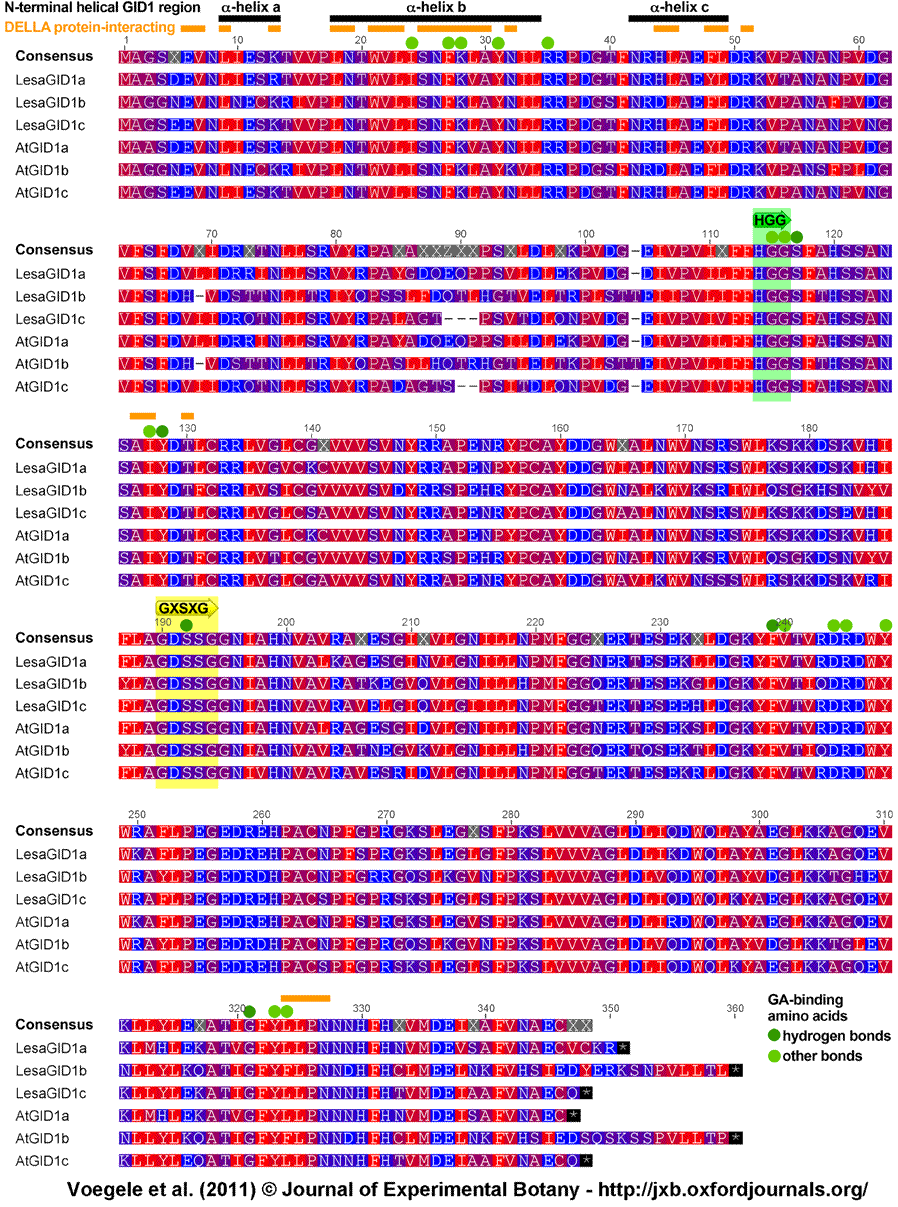
Supplemental Figure S2. Sequence alignment of the predicted GID1 proteins of Lepidium sativum (LesaGID1a, LesaGID1b, LesaGD1c) and Arabidopsis thaliana (AtGID1a, AtGID1b, AtGID1c).
Alignment of LesaGID1a, LesaGID1b, LesaGID1c, AtGID1a, AtGID1b and AtGID1c protein sequences using the BLOSUM62 algorithm (gap open penalty: 12, gap extension penalty: 3). Similarity between the corresponding sequences from L. sativum and Arabidopsis was 96.2% for GID1a, 92.8% for GID1b, and 94.8% for GID1c. Amino acids are color-coded according to their hydrophobic characteristics, with red being most hydrophobic and blue being most hydrophilic. The α-helices a, b and c of the N-terminal helical GID1 region known to be important for DELLA repressor binding (Murase et al., 2008) are shown as black bars at the top. Residues known crystal structures for DELLA repressor binding (Murase et al., 2008) are indicated as orange bars. Green circles indicate important residues for GA3 and GA4 binding based on the crystal structures (Murase et al., 2008); dark green: hydrogen bonds with GA, light green: other bonds with GA. The HGG and GXSXG motives, known as conserved motifs in hormone sensitive lipases, are highlighted as green (HGG) and yellow (GXSXG) boxes and are indicated by arrows. Accession numbers: LesaGID1a, HQ003455; LesaGID1b, HQ003456; LesaGID1c, HQ003457; AtGID1a, At3g05120; AtGID1b, At3g63010; AtGID1c, At5g27320. Based on the similarities and the presence of all the required seqeunce motifs we conclude that the L. sativum proteins are functional GID1 proteins and are encoded by L. sativum gene orthologs of the corresponding Arabidopsis GID1a, GID1b, and GID1c genes.
| Article in PDF format (1.5 MB) Supplementary data file (6.5 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |