Plant Physiology 161: 1903-1917 (2013)
Spatiotemporal seed development analysis provides insight into primary dormancy induction and evolution of the Lepidium DELAY OF GERMINATION1 Genes [W][OA]
School of Biological Sciences, Plant Molecular Science and Centre for Systems and Synthetic Biology, Royal Holloway, University of London, Egham, Surrey TW20 0EX, United Kingdom (KG, AV, GLM);
Web: 'The Seed Biology Place' - www.seedbiology.eu
University of Freiburg, Faculty of Biology, Institute for Biology II, Botany/Plant Physiology, D-79104 Freiburg, Germany (KG, AV, ABM, GLM)
Universität Osnabrück, Fachbereich Biologie, Botanik, D-49069 Osnabrück, Germany (KS, KM)
Received December 24, 2012; Accepted Feburary 19, 2013; Published Feburary 20, 2013.
DOI:10.1104/pp.112.213298
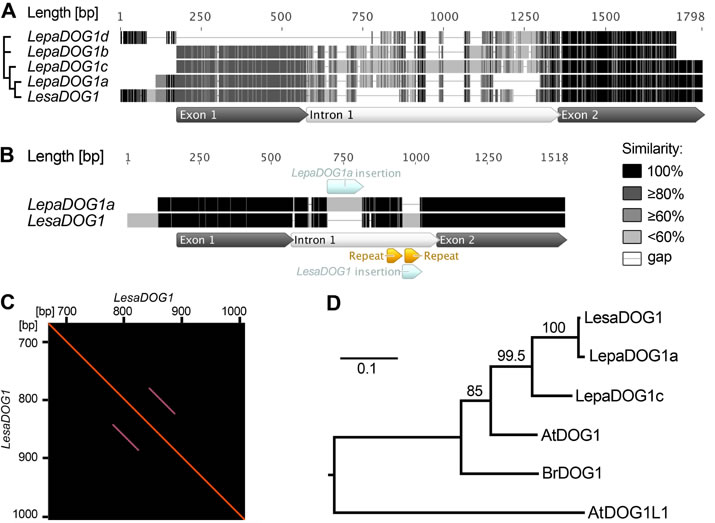
Figure 5. Comparison of Lepidium papillosum (Lepa) and Lepidium sativum (Lesa) DOG1 genomic sequences and phylogenetic analysis of predicted Brassicaceae DOG1 protein sequences.
A, Multiple alignment of the genomic LepaDOG1a (JX512183), LepaDOG1b (JX512184), LepaDOG1c (JX512185), and LepaDOG1d (JX512186) DNA fragments (for details see Supplemental Table S1) as compared to the LesaDOG1 gene (Graeber et al., 2010). We applied a grayscale coloring scheme according to the similarity of the residues in the alignment. A certain residue in a sequence is colored black if it is 100% identical to the other residues in the same alignment column. A residue is colored dark grey if it occurs identically in 80% of the positions in a column (e.g. 4 out of 5 position in a column). A residue is colored moderate gray if it occurs identically in 60% of the positions in a column (e.g. 3 out of 5 positions) or light gray if it occurs identically in less than 60% of the positions in a column (e.g. 1 or 2 out of 5 positions in a column). Internal gaps are treated as unknown characters and end gaps are not considered in the similarity calculation. Gaps are shown as horizontal lines. Annotation of exon-intron structure below the alignment is based on LesaDOG1. Note that the position of the intron splice site is conserved as verified by comparison to LepaDOG1a/c cDNAs. The relationship of the sequences to each other is indicated on the left by a maximum-likelihood cladogram.
B, Pairwise alignment of the genomic sequences of LepaDOG1a with LesaDOG1. Only minor nucleotide changes are present in the two exon regions and the position of the intron with InDels (insertions and deletions) are indicated. Note that in this pairwise alignment only identical residues are colored black, otherwise light grey.
C, Dotblot analysis demonstrating the presence of repeat elements in the LesaDOG1 intron sequence indicated as parallel lines. LesaDOG1 sequence used for this self-dotblot analysis corresponds to that shown in B and the detected repeat locations are marked there. Note that the first repeat sequence is present in LesaDOG1 and LepaDOG1a while the second repeat sequence is missing in LepaDOG1a.
D, Maximum-likelihood phylogenetic tree of predicted protein sequences for Brassicaceae DOG1 genes for which transcript expression in seeds has been demonstrated by Bentsink et al. 2006 (AtDOG1, α-splice variant), Graeber et al. 2010 (LesaDOG1, BrDOG1), and in this work (LepaDOG1a, LepaDOG1c) in relation to the Arabidopsis DOG1-like1 gene (AtDOG1L1). Scale bar indicates substitutions per site, values on branches show bootstrap support. Phylogenetic analysis was performed using PhyML (for details see Materials and Methods).
| Article in PDF format (2 MB) Supplementary data file (4.9 MB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |