Plant Molecular Biology 73: 67-87 (2010)
Cross-species approaches to seed dormancy and germination: conservation and biodiversity of ABA-regulated mechanisms and the Brassicaceae DOG1 genes
Botany / Plant Physiology, Institute for Biology II, Faculty of Biology, University of Freiburg, Schänzlestr. 1, D-79104 Freiburg, Germany, Web: 'The Seed Biology Place' http://www.seedbiology.de (K.G., A.L., K.M., A.R., G.L.-M.)
Department of Cell Biology and Genetics, Palacky University in Olomouc, Slechtitelu 11, 78371 Olomouc-Holice, Czech Republic, and Department of Botany and Plant Physiology, Mendel University of Agriculture and Forestry in Brno, Zemedelska 1, 61300 Brno, Czech Republic (A.W.)
Received August 28, 2009; Accepted November 22, 2009; Published online: December 15, 2009
DOI: 10.1007/s11103-009-9583-x
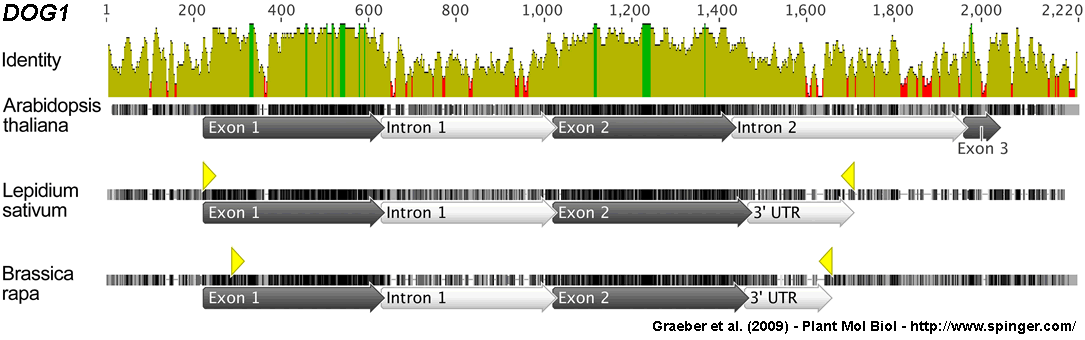
Figure 5. Multiple sequence alignment of Brassicaceae DOG1 (DELAY OF GERMINATION1) gene orthologs. The DOG1 genomic sequences of Arabidopsis thaliana Ler (AtDOG1), Lepidium sativum FR14 (LesaDOG1) and Brassica rapa subsp. pekinensins ‚Chiifu’ (BrDOG1) are compared.
Their GenBank entries and references are: AtDOG1 (EF028470.1, Bentsink et al. 2006), LesaDOG1 (GQ411193, this work) and BrDOG1 (AC189537.2, genomic clone KBrH003E13 from bp 8949 to bp 10992). The identity plot and the alignment of the individual genomic DNA sequences are presented. The exon/intron annotation is based on the Arabidopsis genomic DNA sequence for AtDOG1. For Arabidopsis splicing of the introns as they are drawn here generates the known AtDOG1 alpha-type transcripts (Bentsink et al. 2006). To verify splice sites and to identify 3' UTR regions we cloned LesaDOG1 and BrDOG1 cDNAs: the yellow triangles mark the verified cDNA sequence regions and their GenBank entry numbers are QG411192 (LesaDOG1) and QG411194 (BrDOG1). The identity plot displays the average identity across all genomic sequences for every position in a 10 bp window. High green bars indicate that the residues at the position of the window are the same across all sequences. Medium yellow bars represent less than complete identity and red low bars refer to very low identity for the given window position. Thick horizontal lines represent the individual genomic DNA sequences, thin horizontal lines represent gaps in the alignment. Different vertical shadings represent different levels of identity at this base position between the three sequences: black = all bases at this position are identical in all three sequences, dark grey= two of the three sequences have the same base at this position, light grey = this base at this position is only present in this sequence.
| Article in PDF format (872 KB) Abstract of PMB 2009 |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |