The Plant Cell 23: 2045-2063 (2011)
A Guideline to Family-wide Comparative State-of-the-art qRT-PCR Analysis Exemplified with a Brassicaceae Cross-species Seed Germination Case Study [W][OA]
* Both authors contributed equally to this work
The University of Nottingham, Division of Statistics, School of Mathematical Sciences, University Park, Nottingham NG7 2RD, United Kingdom (A.T.A.W.)
Received February 8, 2011; revised May 6, 2011; accepted May 27, 2011; published June 10, 2011.
www.plantcell.org/cgi/doi/10.1105/tpc.111.084103
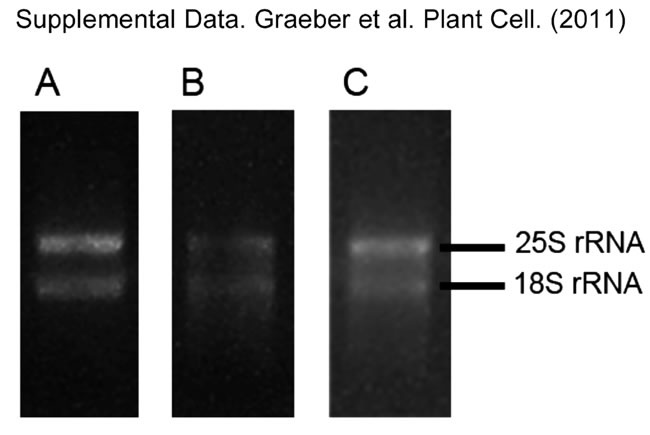 |
Supplemental Figure 1. Determination of RNA integrity via gel electrophoresis. (A) RNA with high integrity used for further analyses: to clear bands for the 25S and 18S rRNA, with the upper band being twice as strong as the lower one. (B) and (C) RNA with low integrity, which was not further used: showing either not the 2:1 ratio in band intensity between the 25S and the 18S rRNA (B) or showing RNA degradation indicated by a smear on the gel (C). Between 100 and 200ng RNA were loaded on a 1% agarose gel. |
Synopsis: Developmental processes like seed germination are characterised by massive transcriptome changes. This study compares seed transcriptome datasets of different Brassicaceae to identify stable expressed reference genes for cross-species qRT-PCR normalisation. A workflow is presented for improving RNA quality, qRT-PCR performance, and normalisation when analysing expression changes across species.
| Article in PDF format (1.8 MB) Supplemental data file (156 KB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |