The Plant Cell 23: 2045-2063 (2011)
A Guideline to Family-wide Comparative State-of-the-art qRT-PCR Analysis Exemplified with a Brassicaceae Cross-species Seed Germination Case Study [W][OA]
* Both authors contributed equally to this work
The University of Nottingham, Division of Statistics, School of Mathematical Sciences, University Park, Nottingham NG7 2RD, United Kingdom (A.T.A.W.)
Received February 8, 2011; revised May 6, 2011; accepted May 27, 2011; published June 10, 2011.
www.plantcell.org/cgi/doi/10.1105/tpc.111.084103
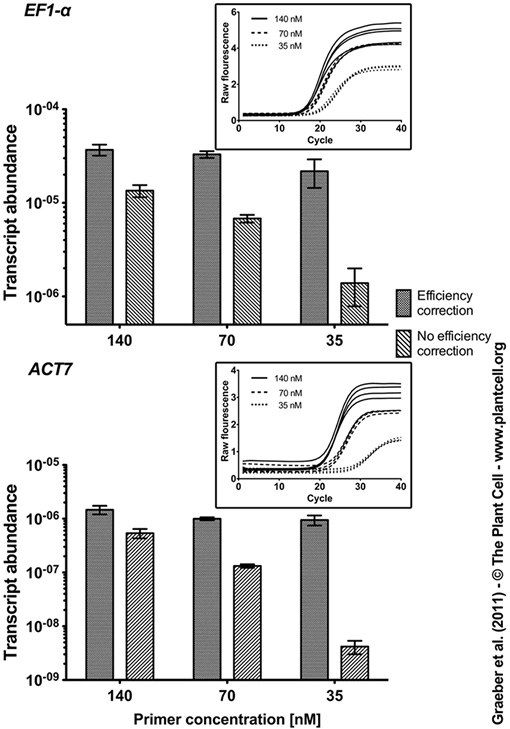 |
Figure 1. Effect of Different PCR Efficiencies on the Measured Transcript Abundance of EF1-α and ACT7 with and without Efficiency Correction. Different PCR efficiencies were obtained by varying the concentrations of the gene-specific primers as described in Table 2. Lowered primer concentration results in reduction of PCR efficiency recognizable by decreased steepness of the exponential phases in the qRT-PCR fluorescence curves (insets in the upper right corners; shown are individual amplification plots for 3 different primer concentrations, 4 biological replicates each). PCR efficiencies were determined by PCR Miner software. Efficiency-corrected transcript abundance was calculated as describe in methods by using the average efficiency of all samples for each gene and each primer concentration. Non-efficiency corrected transcript abundance was calculated from the CT values determined by PCR Miner assuming 100 % PCR efficiency. Mean values ± SD of 4 biological replicates are presented. |
Synopsis: Developmental processes like seed germination are characterised by massive transcriptome changes. This study compares seed transcriptome datasets of different Brassicaceae to identify stable expressed reference genes for cross-species qRT-PCR normalisation. A workflow is presented for improving RNA quality, qRT-PCR performance, and normalisation when analysing expression changes across species.
| Article in PDF format (1.2 MB) Supplemental data file (156 KB) |
|
|
|
The Seed Biology Place |
Webdesign Gerhard Leubner 2000 |